Winter 2022
|
A follow up to our review (below) on the importance of evolutionary potential to extinction risk has been published in the Journal of Heredity! This open access simulation study of the endangered southwestern willow flycatcher illustrates how evolutionary potential interacts with dispersal capacity, demography, and environmental stochasticity to impact population trajectories under climate change. This research provides a framework for integrating our increasing insights into evolutionary potential into extinction risk assessments and recovery planning for threatened and endangered species. |
|
Our open access review of evolutionary potential and extinction risk is on the cover of November's Frontiers in Ecology and the Environment! Evolutionary potential can reduce a species’ extinction risk by facilitating adaptive responses to environmental change. It is a critical component of a species’ adaptive capacity yet is rarely included in extinction risk assessments. This is largely because evolutionary potential is challenging to evaluate for species of conservation concern, due to its complexity and multidimensionality. However, there are a number of proxies for evolutionary potential (reviewed in our paper) that can be estimated from environmental, phenotypic, and genetic data. Including evolutionary potential in extinction risk assessments is rare. The best available models (reviewed in our paper) integrate demographic and evolutionary dynamics with environmental change, though these models will be challenging to parameterize for data-deficient species. Where data are limited, best practices for maintaining evolutionary potential remain essential: conserving across the breadth of adaptive diversity and protecting the integrity of processes that drive evolutionary change. Integration of evolutionary potential into decision-making will improve extinction risk assessments & conservation planning to ensure resilience in the face of complex change. This integration is challenging but essential & remains an important area for innovation in applied conservation science.
|
Fall 2022
|
Our open access paper on delineating conservation units is out in Molecular Ecology! We illustrate how a carefully planned genetic study, designed to address priority management goals that include conservation unit delineation, can provide multiple insights to inform conservation action. Columbia spotted frogs occupy fragmented aquatic habitats in Nevada, a region that has seen substantial anthropogenic changes over the past 200 years. Invasive species, disease, drought, and climate change place many populations at high risk of extirpation without management intervention. Long a candidate for listing under the ESA, these frogs were removed from the candidate list in 2015 due to proactive conservation actions, however many populations continued to decline. A genetic study was initiated to inform conservation unit delineation and prioritize conservation actions. The large number of occupied sites required a careful study design to maximize competing management goals. Combined with data on dispersal and occupancy + landscape predictors, we used a suite of approaches to evaluate and reassign conservation units, and assess connectivity and neutral + adaptive diversity. In particular, using gravity models predicted across occupied but (genetically) unsampled sites, we found unexpected patterns of functional connectivity that highlighted the uniqueness of these desert populations and better informed the reassignment of management units. Most populations have extremely low genetic diversity and effective population sizes; many are also at risk of extirpation from bullfrogs. Our results are informing genetic rescue, translocation, and recovery efforts targeted to maintain genetic distinctiveness while boosting genetic diversity.
|
Summer 2022
|
We've posted our review of the past, present, and future of genetic and genomic data in U.S. Endangered Species Act listing and recovery work. This is a book chapter written by myself and Dr. Tanya Lama for a forthcoming book celebrating the 50th anniversary of the ESA. We review how genetic data have informed decision-making under the ESA, and how the transition to genomics is improving the information that we can apply to both listing and recovery decisions. In some cases, genomic data are presenting new challenges to applied conservation under the ESA, providing an opportunity to evaluate and innovate existing practices. In all cases, falling costs and the increasing ease of genomic-scale data production in at-risk species are providing an unparalleled opportunity to improve applied conservation of threatened and endangered species. These advances are expanding new frontiers for agency use of the “best available science” in ESA implementation. Our chapter will be published in 2023 in The Codex of the Endangered Species Act: The Next Fifty Years - Volume II, edited by Lowell E. Baier and John Organ.
|
Fall 2021
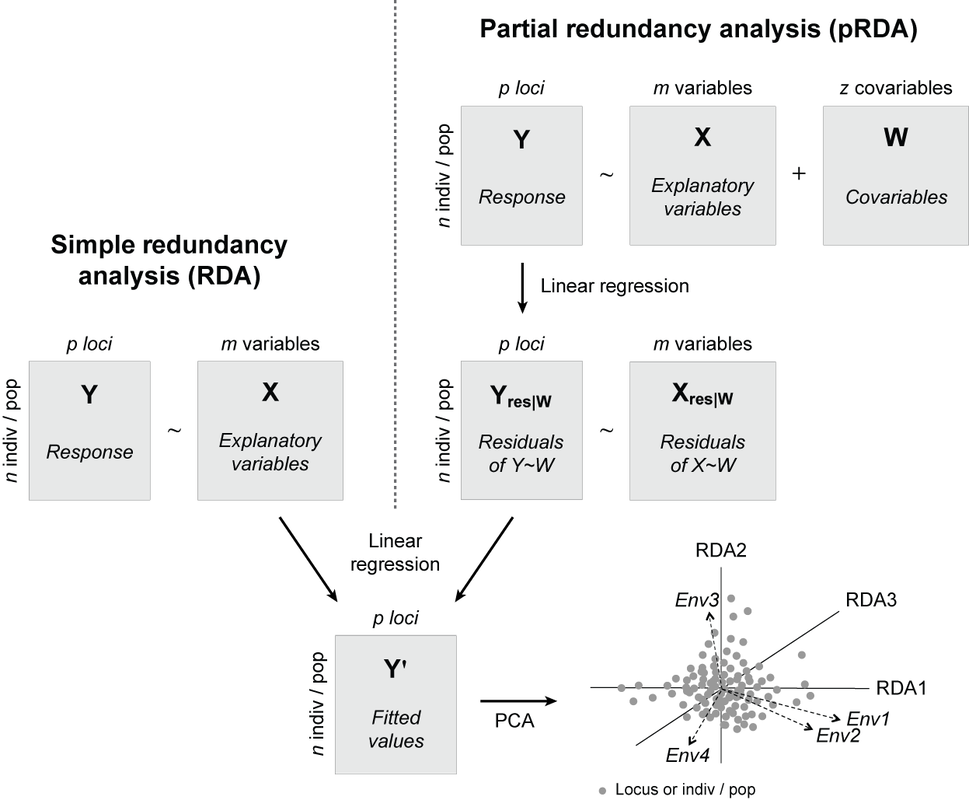
A comparison of procedures for fitting a simple RDA (left) and a partial RDA (right). Both share a common last step of computing a principal components analysis (PCA) of the fitted values, Y′. An example RDA biplot is shown, where RDA1, RDA2, and RDA3 are the orthogonal canonical axes, the dashed vectors represent the environmental predictors, and the grey points represent either the loci or individuals/populations (depending on which the user chooses to represent). The arrangement of these items in the ordination space reflects their relationship with the canonical axes. Figure inspired by Legendre & Legendre (2012).
|
Our open access review on redundancy analysis (RDA) for landscape genomics is now available in Methods in Ecology and Evolution! This was a team effort between myself and my amazing colleague Thibaut Capblancq. The goals of our review are to provide resources to the landscape genomics community to improve understanding of RDA as a modelling framework, and encourage its appropriate use across diverse landscape genomics applications.
Using a large lodgepole pine data set, we explore the many applications of RDA, including variable selection, variance partitioning, genotype-environment association testing, and the calculation of adaptive indices and genomic offset. We illustrate the use of RDA to address four of the major questions in landscape genomics:
We've provided a user-friendly R tutorial that includes code, explanation, and interpretation for all analyses included in the paper. We hope the review will be useful to the landscape genomics community! |
Summer 2021
Job news! I will be joining the new Branch of SSA Science Support with the U.S. Fish and Wildlife Service in October! The Species Status Assessment (SSA) framework delivers foundational science for informing U.S. Endangered Species Act decision-making. I'll be providing analytical support for the development of robust, scientifically-rigorous SSAs, and framework support to improve integration of genomic data into decision-making.
This is a dream job for me, allowing me to extend my Smith Fellowship research on effective practices for incorporating genomic data into ESA listing and recovery decision-making. I'm thrilled to be applying my skills to conservation research and applications that support the protection of biodiversity during this critical period of unprecedented biodiversity loss.
This is a dream job for me, allowing me to extend my Smith Fellowship research on effective practices for incorporating genomic data into ESA listing and recovery decision-making. I'm thrilled to be applying my skills to conservation research and applications that support the protection of biodiversity during this critical period of unprecedented biodiversity loss.
Spring 2021
|
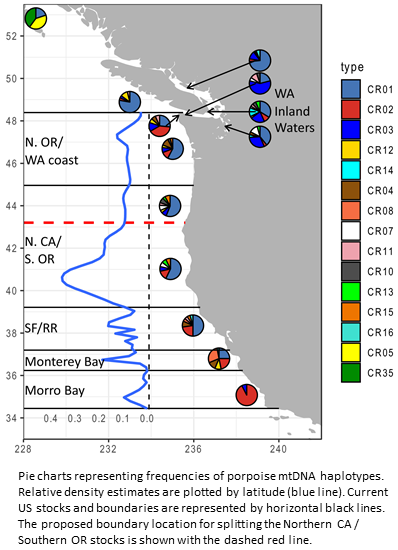
We have a new paper published in Molecular Ecology on the North Pacific harbor porpoise, a nearshore species that is very difficult to sample in the wild. In this study, led by Phillip Morin at the Southwest Fisheries Science Center, we use microhaplotype data derived from opportunistic and degraded tissue samples to examine genetic structure in this species from British Columbia to California.
We identified strong isolation by distance, suggesting restricted dispersal, along with regions of current or historic barriers to gene flow. In particular, the southernmost population in California is genetically distinct, with reduced genetic variability and an otherwise rare mtDNA haplotype. Southern populations also show signatures of potential local adaptation related to sea surface height and temperature. These results will inform coastal management stock delineations and other future conservation efforts. |
Winter 2020
|
I am pleased to take on a leadership role with the Society for Conservation Biology Conservation Genetics Working Group, as new Committee Chair of the CGWG Strategic Planning Committee.
As part of that role, I am leading an initiative to develop a database linking conservation practitioners with genomics experts to facilitate productive integration of genetic and genomic data into conservation and management decision-making. You can join this effort here, by adding your information to the spreadsheet linked at the bottom of the page. The new CGWG logo was designed by Dr. Stephanie Galla at Boise State University. The design features a helix motif in a radial pattern, reflecting how DNA is used to understand demographic patterns in nature. Within these helices are a subtle rainbow design, echoing our commitment to preserving genetic diversity, and also keeping diverse researchers and practitioners at the table. Within this design, people may be reminded of several species, or elements of nature — for example, a flower, a sea urchin, or a jellyfish. This broad pattern is meant to embody diverse lifeforms, which we aim to preserve through the use of molecular tools. |
Spring 2020
|
We have a new preprint available, and the first manuscript from our Team Ascaphus collaboration! Co-led by former undergraduate researcher and now B.Sc. degree-holder Christina Martinez and Ph.D. student Amanda Cicchino, we used an integrative eco-evo-devo framework to test the direct and indirect effects of developmental processes and environmental selection on labial tooth number in coastal and Rocky Mountain tailed frog populations.
Christina counted A LOT of tadpole teeth (over 150,000!), and all of that counting helped us understand how tadpole mouthpart morphology varies not only between species, but also across sites, and in response to variation in local environmental conditions and developmental processes. Excellent research by one of the all-star undergraduate researchers on our Ascaphus Team. Congratulations Christina! |
Winter 2019
I have been selected as a David H. Smith Conservation Research Fellow! I'll be investigating how best to integrate genomic data into listing practices under the U.S. Endangered Species Act. The Smith Fellows program gives me the opportunity to work closely with a group of academic and conservation practitioner mentors to ensure that conservation science is effectively linked to conservation actions. It really couldn't be a better fit for my goals as a conservation scientist.
I'm looking forward to continuing research that has a real impact on conserving biodiversity and preventing extinction during our current period of rapid, massive, and unprecedented global change.
I'm looking forward to continuing research that has a real impact on conserving biodiversity and preventing extinction during our current period of rapid, massive, and unprecedented global change.
|
The latest update for CDPOP & CDMetaPOP is now available! This module provides a flexible framework for simulating multilocus selection so users can evaluate interactions among gene flow, demography, and complex selection-driven processes across multivariate landscapes.
Our manuscript includes a box that explains the evolution of CDPOP (Cost-Distance POPulations) & CDMetaPOP (Cost-Distance Meta‐POPulations), including features in common and unique to each simulation platform. This will be helpful to both new and existing CDPOP & CDMetaPOP users. This update was a collaborative effort with Erin Landguth, Andrew Eckert, Andrew Shirk, Mitra Menon, Amy Whipple, Casey Day, and Sam Cushman. See a nice interview with Erin about the project here. |
Summer 2019
|
It has been a busy summer of workshops and symposia!
|
Spring 2019
|
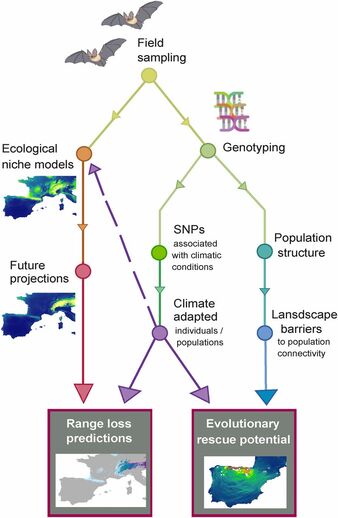
I'm thrilled to announce the publication of our integrative paper led by Orly Razgour & now available in PNAS: Considering adaptive genetic variation in climate change vulnerability assessment reduces species range loss projections.
See a nice summary and interview with Orly about the paper in Wired Magazine. Forecasts of species vulnerability and extinction risk under future climate change commonly ignore local adaptations despite their importance for determining the potential of populations to respond to future changes. We present an approach to assess the impacts of global climate change on biodiversity that takes into account adaptive genetic variation and evolutionary potential. Focusing on two Mediterranean bat species, we show that considering local climatic adaptations reduces range loss projections but increases the potential for competition between species. Our findings suggest that failure to account for within-species variability can result in overestimation of future biodiversity losses. Therefore, it is important to identify the climate-adaptive potential of populations and to increase landscape connectivity between populations to enable the spread of adaptive genetic variation. |
|
April brought a trip to the CSIRO Lab in Hobart, Tasmania, where I taught landscape genomics at the Population Genomics in R workshop. What a fantastic group of co-instructors and students! Thanks to the CSIRO Environomics Future Science Platform for sponsoring the workshop.
The 2019 US-IALE meeting (International Association for Landscape Ecology) was held in Fort Collins, CO on April 7-11, with the theme of Conservation Innovation. IALE included two landscape genetic symposiums this year, including a special session hosted by myself and Melanie Murphy: "Conservation innovation using landscape genetics: novel approaches of integrating landscape ecology and population genetics to address conservation problems." |
Fall 2018
Our new book chapter "Landscape Genomics for Wildlife Research" is now published, in collaboration with Erin Landguth, Brian Hand, and Niko Balkenhol. We review the use of adaptive landscape genomics in applied wildlife research, and discuss its untapped potential to inform management and conservation decision-making. In particular, we emphasize the need to move beyond correlative genotype-environment association tests, and discuss the value of integrating complementary data and analytical methods to improve our understanding of adaptation in wildlife species.
NSF has funded our “Rules of Life” EAGER grant entitled “Landscape Phenomics: Predicting vulnerability to climate change by linking environmental heterogeneity to genetic and phenotypic variation”. Our goal is to predict which populations are most vulnerable to environmental change by first understanding how environmental variation shapes genetic and phenotypic variation in resilience traits. We are focusing on tailed frogs (Ascaphus spp.) for this research since they are sensitive to high temperatures, play an important role as grazers in streams, and are amenable to the genomic and physiological work we are doing. Our fantastic team for this research includes: Amanda Cicchino, Erin Landguth, Jason Dunham, Chris Funk, and Cameron Ghalambor.
NSF has funded our “Rules of Life” EAGER grant entitled “Landscape Phenomics: Predicting vulnerability to climate change by linking environmental heterogeneity to genetic and phenotypic variation”. Our goal is to predict which populations are most vulnerable to environmental change by first understanding how environmental variation shapes genetic and phenotypic variation in resilience traits. We are focusing on tailed frogs (Ascaphus spp.) for this research since they are sensitive to high temperatures, play an important role as grazers in streams, and are amenable to the genomic and physiological work we are doing. Our fantastic team for this research includes: Amanda Cicchino, Erin Landguth, Jason Dunham, Chris Funk, and Cameron Ghalambor.
|
Our paper "Improving conservation policy with genomics: a guide to integrating adaptive potential into U.S. Endangered Species Act decisions for conservation practitioners and geneticists" is now out in Conservation Genetics. This review was a collaborative effort between Chris Funk and myself at Colorado State University, Sarah Converse with USGS/WA Cooperative Fish and Wildlife/University of Washington, Cat Darst with US Fish and Wildlife (Ventura, CA), and Steve Morey with US Fish and Wildlife (Portland, OR).
|
Summer 2018
|
Amanda Cicchino (CSU PhD student), Katherine Pain (CSU undergraduate), and I kicked off a productive Ascaphus field season in Oregon this June. Amanda and Kat are continuing our physiological and genomics work with Coastal Tailed Frogs (A. truei) in Oregon through most of the summer.
June also sees publication of our ConGen 2017 meeting review led by Sarah Hendricks: "Recent advances in conservation and population genomics data analysis". This comprehensive review of the week-long meeting includes discussion of: increasing the participation of women in population and conservation genomics; the need for data visualization and its importance in finding problematic data; the effects of data filtering choices on downstream analyses; and the increasing availability of whole‐genome sequencing and the new challenges these data present. |
Spring 2018
Our paper "Comparing methods for detecting multilocus adaptation with multivariate genotype-environment associations" has been accepted in Molecular Ecology. We evaluate four multivariate GEA methods and five univariate and differentiation-based approaches, using simulations of multilocus selection. We found that constrained ordinations, particularly redundancy analysis (RDA), showed a superior combination of low false positive and high true positive rates across all levels of selection. We illustrate the use of RDA by reanalyzing genomic data from North American wolves, highlighting the unique, covarying sets of adaptive loci that can be identified using this multivariate genotype-environment association method.
Fall 2017
|
Tens of thousands of flipped logs and 5,083 red-backed salamanders later, results from our resurvey of 50 sites across New England for red-backed salamander color polymorphism is now available in Ecography! Check it out here.
Our new paper on genomics and conservation is now out in Evolutionary Applications! This collaboration came out of the NIMBioS Investigate Workshop on Next Generation Genetic Monitoring that was held in Knoxville, TN in November of 2016. Looking forward to seeing contributions from other groups at the workshop as part of this special issue of Evolutionary Applications. |
Summer 2017
I've moved to colorful Colorado to start a postdoc in Chris Funk's lab at Colorado State University. I will be working on the population genomics and evolutionary ecology of coastal tailed frogs (Ascaphus truei) and Rocky Mountain tailed frogs (A. montanus). I'm really looking forward to working with this dynamic and accomplished group of scientists!
I'll also be teaching landscape genomics at ConGen 2017 in September at beautiful Flathead Lake. Thanks to the organizers for the invitation - it should be a great course! |
Spring 2017
I have completed my PhD at Duke University! Many thanks to my advisor, Dean Urban, my committee, and my lab mates for their support over the years. And a huge thank you to my talented colleague and wonderful friend Danica Schaffer-Smith for making this amazing design for our latest Landscape Ecology Lab t-shirt!
|
Fall 2016
Helene Wagner, Mariana Chávez-Pesqueira, and I have published a new outlier detection method in Molecular Ecology Resources. This method uses Moran eigenvector maps for spatial detection of outlier loci. You can find the associated tutorial and vignette here.
Kemen Austin has an important new publication in Environmental Research Letters which estimates the extent and location of suitable land for oil palm cultivation in Gabon, in an effort to minimize negative environmental impacts. Kemen worked on these models in the Species Distribution Modeling class I taught at Duke in 2015, and I am proud to a contributor to this important publication: "An assessment of High Carbon Stock and High Conservation Value approaches to sustainable oil palm cultivation in Gabon".
Kemen Austin has an important new publication in Environmental Research Letters which estimates the extent and location of suitable land for oil palm cultivation in Gabon, in an effort to minimize negative environmental impacts. Kemen worked on these models in the Species Distribution Modeling class I taught at Duke in 2015, and I am proud to a contributor to this important publication: "An assessment of High Carbon Stock and High Conservation Value approaches to sustainable oil palm cultivation in Gabon".
|
Last day of mountain field work with my wonderful collaborators and friends Lori Williams & Charles Lawson, with birder extraordinaire Clifton Avery along as well (all with the NC Wildlife Resources Commission). A lovely day sampling Weller's Salamanders at Sugar Mt, NC, with a stop on the Blue Ridge Parkway on the way home. Thanks to this amazing team for all their help! |
Summer 2016
|
Summer is a great time to collect baseline data on disease prevalence in the mountain salamanders of western North Carolina. In this photo, a red eft is being swabbed by my collaborator Lori Williams (NC Wildlife Resources Commission). We will test the sample for the fungal pathogens Bd and Bsal. This work is funded by our grant from the Triangle Center for Evolutionary Medicine.
I am honored to be funded in the last year of my dissertation research by PEO International, a philanthropic organization devoted to celebrating the advancement of women, and funding education for women to help them achieve their highest aspirations. Thank you to my local chapter for sponsoring my application! |
Spring 2016
|
The Triangle Center for Evolutionary Medicine has funded our proposal on "Emerging human-mediated pathogens in North Carolina amphibians and reptiles". I am thrilled to be a co-PI (with colleagues from NCSU, NCCU, the NC Museum of Natural Sciences, and the NC Wildlife Resources Commission) on this very important project investigating the distribution and impact of four emerging wildlife pathogens in North Carolina. These fungal and viral infections have been associated with morbidity, mortality, and mass die-offs of wild amphibians and reptiles around the world.
Our new paper, "Detecting spatial genetic signatures of local adaptation in heterogeneous landscapes", is now available! It has been published as part of the Molecular Ecology special issue on "Detecting selection in natural populations: making sense of genome scans and towards alternative solutions." |
Fall 2016
|
I am the proud recipient of a Katherine Goodman Stern Fellowship, which supports advanced graduate students in the write-up stage of their dissertation. Thanks to the Katherine Stern endowment for this award!
The American Society of Naturalists, who must surely have one of the most fantastic logos of all time, has awarded me a Student Research Fellowship to support my work on local adaptation in Weller's Salamander. |
Spring 2015
I was delighted to return to my M.Sc. alma mater, Huxley College at Western Washington University, as part of the Huxley Speaker Series. I gave a talk on "Genomics and Conservation; or, Salamanders vs. Climate Change".
I have been awarded a National Science Foundation Doctoral Dissertation Improvement Grant to fund my research on local adaptation in red-backed salamanders living in urban heat islands.
My invited article on "Mountains, Climate Change, and Conservation: The Role of Landscape Genomics" is now available in Mountain Views, the newsletter of the Consortium for Integrated Climate Research in Western Mountains.
I have been awarded a National Science Foundation Doctoral Dissertation Improvement Grant to fund my research on local adaptation in red-backed salamanders living in urban heat islands.
My invited article on "Mountains, Climate Change, and Conservation: The Role of Landscape Genomics" is now available in Mountain Views, the newsletter of the Consortium for Integrated Climate Research in Western Mountains.
Fall 2014
|
I was honored to give an invited talk at this year's MtnClim Conference in Midway, Utah. I spoke to an incredibly diverse group of climatologists, paleoecologists, sociologists, and biologists on conservation applications of landscape genomics in mountain systems under climate change. We also took a fantastic field trip to the beautiful and unusual (east-west running) Uinta Mountains.
My colleagues at Western Washington University and I have a new paper in Ecology and Evolution on speciation in the arctic-alpine plant genus Campanula. I gave an invited talk at the Fall 2014 meeting of the North Carolina Herpetological Society. I spoke about my dissertation research on the landscape genomics of Weller's Salamander in the southern Appalachians. I gave an invited seminar at the Fall Biology Seminar Series at North Carolina Central University on applications of landscape genomics in species of conservation concern. |